Thème de recherche n°2 : Méthylation de l'appareil traductionnel
The translation apparatus (ribosome, translation factors and transfer RNAs) is the target of multiple post-transcriptional and post-translational modifications. Methylation is one of the most common modifications, and a significant percentage of proteins, across all organisms, catalyze the transfer of a methyl group from S-adenosyl-methionine (SAM) to a substrate. In humans, one third of these methyltransferases (MTase) have been linked to cancer and mental disorders. However, little is known about the function of these modifications.
We want to understand of the biological functions of methylation of RNA and proteins involved in translation. Remarkably, in yeast and humans, two thirds of known MTases are dedicated to the translation machinery. We are interested in Trm112, a yeast protein placed at the interface between ribosome synthesis and translation, which functions as an activating platform of four methyltransferases (MTases) with different substrates. Our goal is to gain insights into the function of the MTases interacting with Trm112.
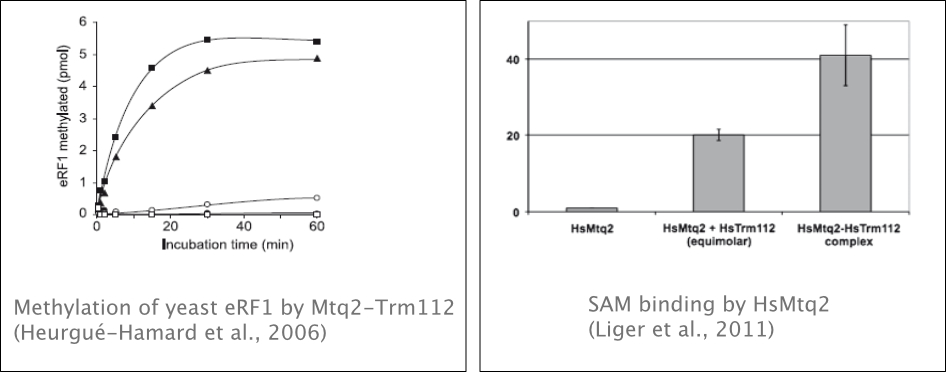
We previously showed that class 1 translation termination factors were methylated in bacteria (E. coli, C. trachomatis) and eukaryotes (S. cerevisiae, Homo sapiens, Mus musculus). These proteins are essential for releasing a functional protein when the ribosome encounters a stop codon on the mRNA. Methylation occurs on the glutamine from the conserved Gly-Gly-Gln motif, which is involved in catalyzing peptide release at the ribosomal peptidyl-transferase center. We identified the methyltransferases (MTases) involved in this process, characterized their function, and determined their structure (for a review, see Graille et al., 2012). By using genetic and biochemical approaches, we characterized the molecular mechanism of these MTases and the activating function of Trm112 (Heurgué-Hamard et al., 2006, Liger et al., 2011).
Trm112 interacts with four MTases involved in translational apparatus modification: Mtq2 that methylates translation termination eRF1, Trm9 and Trm11 that methylate tRNAs and Bud23 that methylates 18S ribosomal RNA. We recently showed that Bud23-Trm112 complex methylates specifically G1575 base in 18S rRNA. Trm112 interacts directly with Bud23 in vitro, and it is required for its stability in vivo. Consequently, in the absence of Trm112, Bud23 is no longer able to bind nascent pre-ribosomes, leading to 40S subunit defect. Bud23–Trm112 also forms a ternary complex with the DEAH Box Helicase Dhr1. This helicase contributes to efficient recruitment of Bud23–Trm112 to pre-ribosomes (Figaro
et al., 2012., Létoquart et al., 2014). We also showed that Trm112 is involved in 60S subunit synthesis through its interaction and stabilization of Mtq2. This MTase is part of nuclear pre-60S subunits, being required for the efficient production of 5.8S and 25S rRNAs and subunit export. Mtq2 may play a role in surveilling the formation of functional and essential ribosomal sites (Lacoux et al., 2020).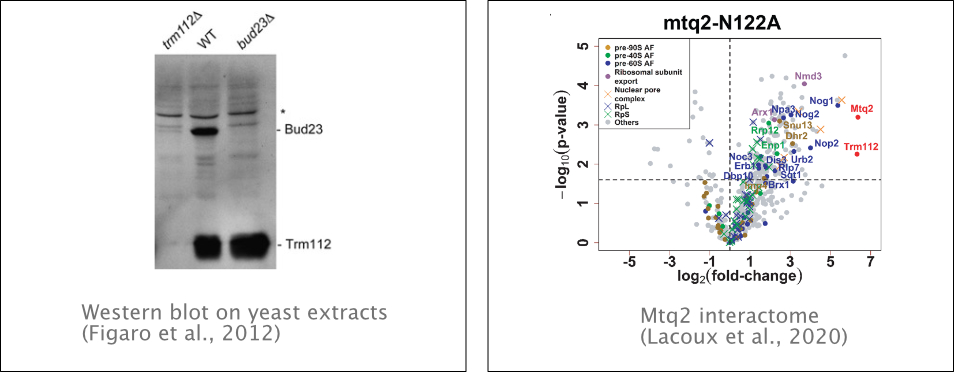
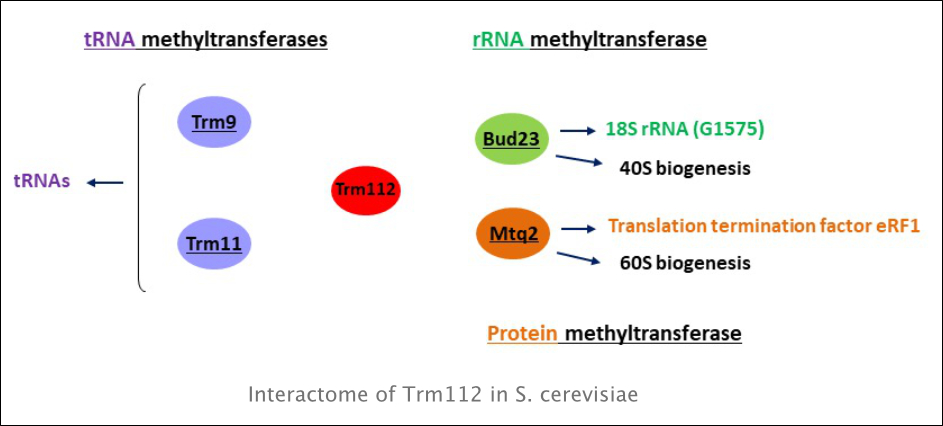
Trm112 serves as an activating platform for four methyltransferases (MTases) at the interface between ribosome synthesis and function to potentially regulate both processes. We would like to decipher at molecular level the role of these methylations in translation, cell biology in general and the link with diverse pathologies. Our experiments are mainly done in yeast S. cerevisiae, using complementary experimental approaches such as yeast genetics, molecular biology, physiological studies, biochemistry, immunoprecipitation followed by mass spectrometry.
Collaborators: Dr Marc Graille (Ecole Polytechnique, Palaiseau, France), Pr Denis L.J. Lafontaine (Université Libre de Bruxelles, Charleroi, Belgium), Dr Sarah Cianférani and Dr Christine Carapito (Institut Pluridisciplinaire Hubert Curien, Strasbourg, France)
